
Tools and Strategies for Long-Read Sequencing and De Novo Assembly of Plant Genomes: Trends in Plant Science

Sequencing from scratch: reference genomes and de novo sequence assembly – HudsonAlpha Institute for Biotechnology

Sequencing from scratch: reference genomes and de novo sequence assembly – HudsonAlpha Institute for Biotechnology

Long walk to genomics: History and current approaches to genome sequencing and assembly - ScienceDirect
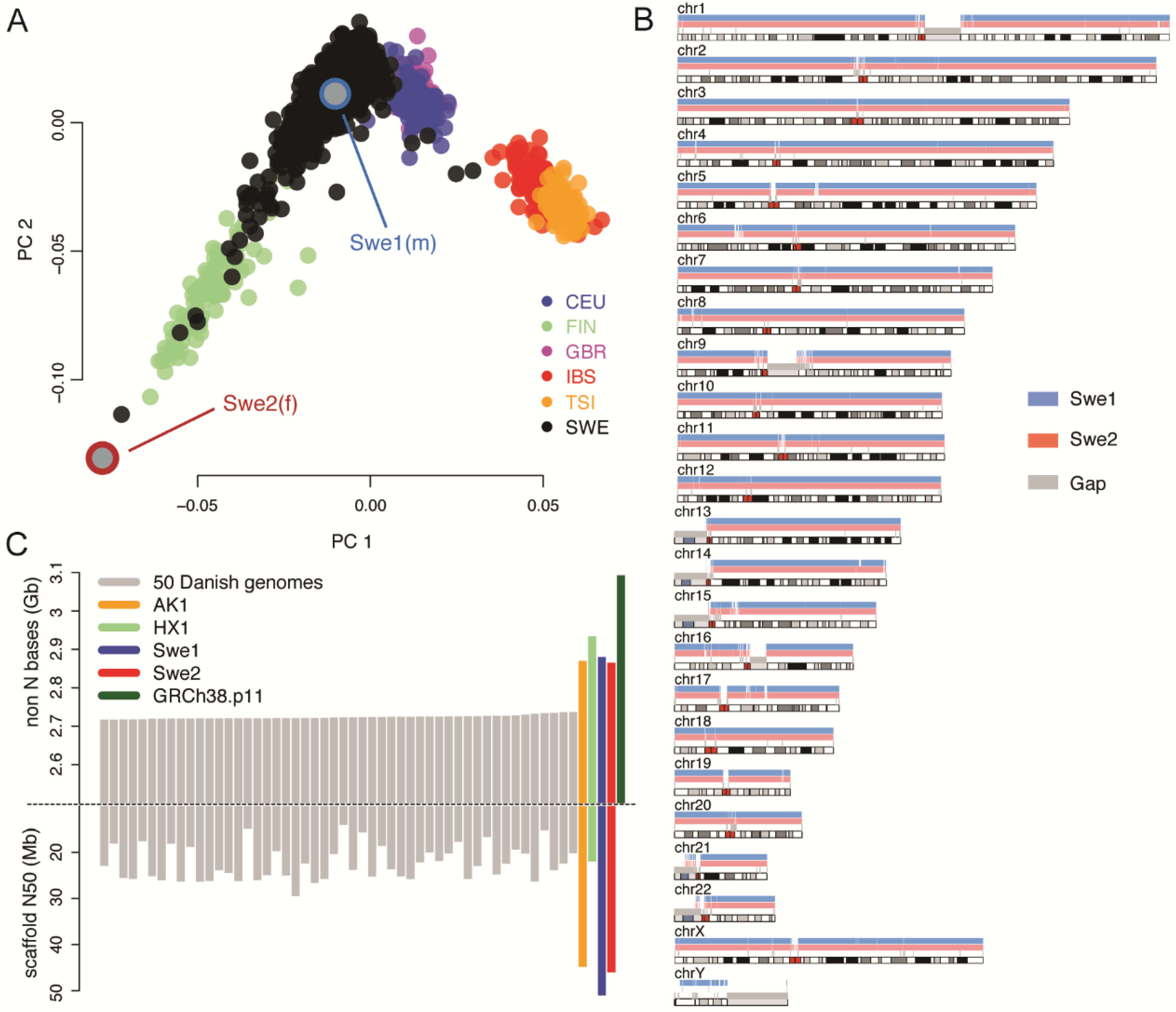
Genes | Free Full-Text | De Novo Assembly of Two Swedish Genomes Reveals Missing Segments from the Human GRCh38 Reference and Improves Variant Calling of Population-Scale Sequencing Data

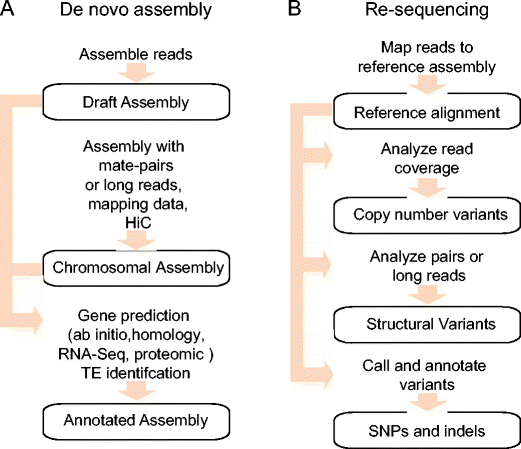
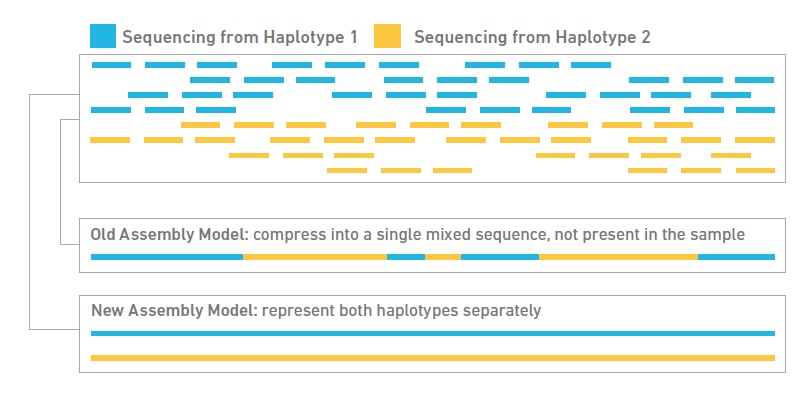


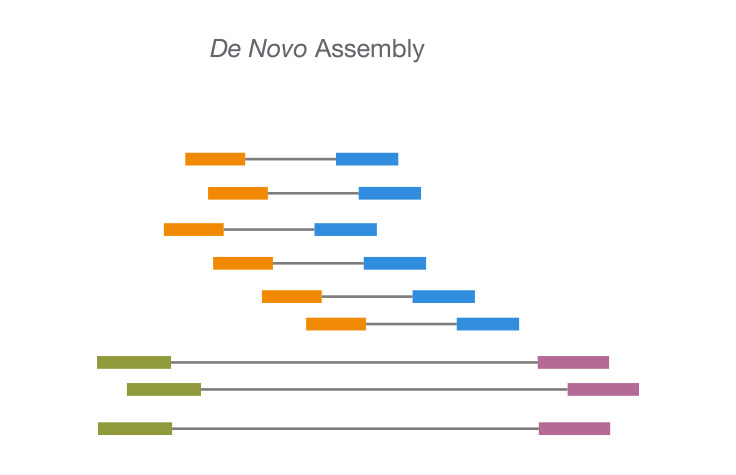
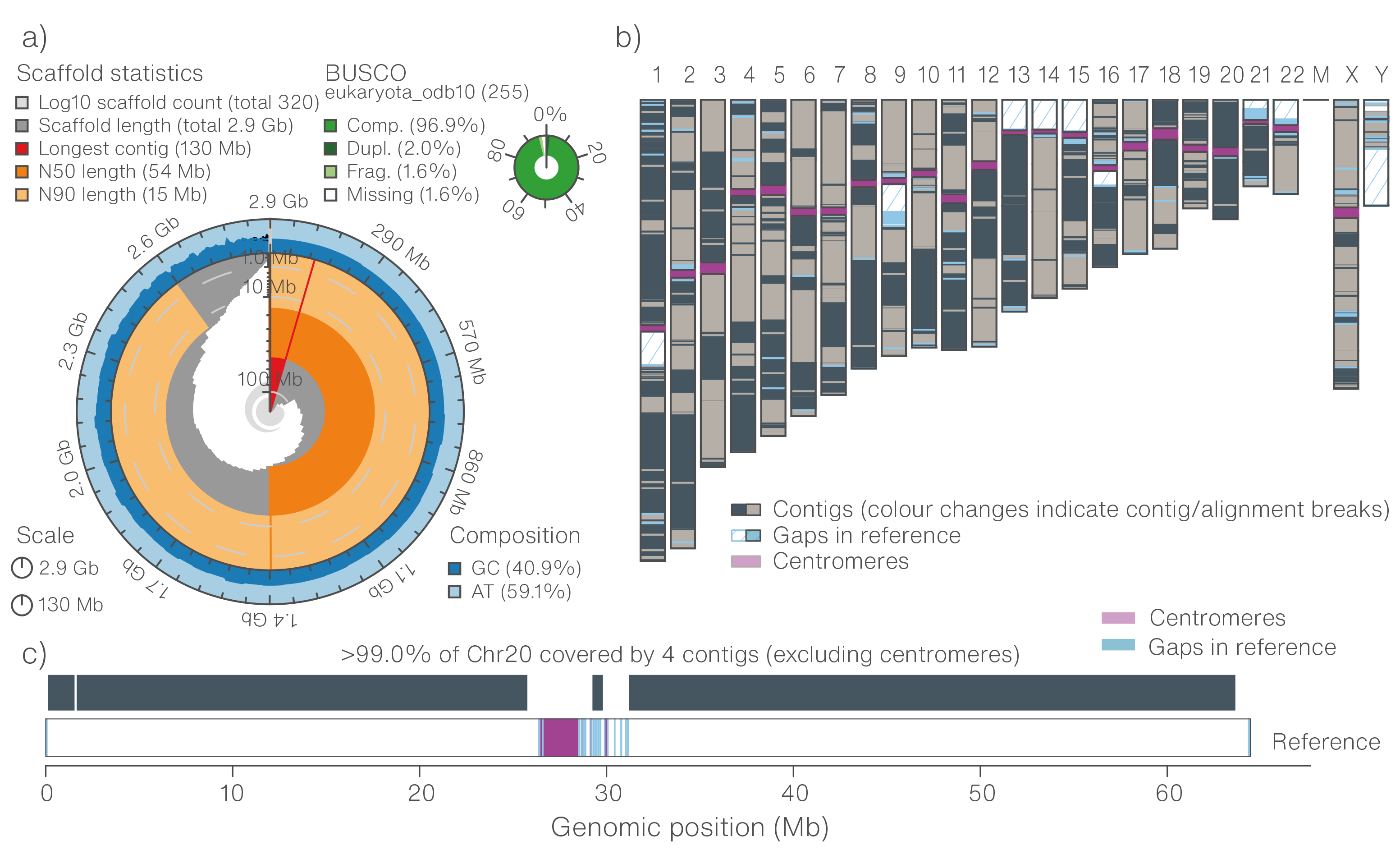


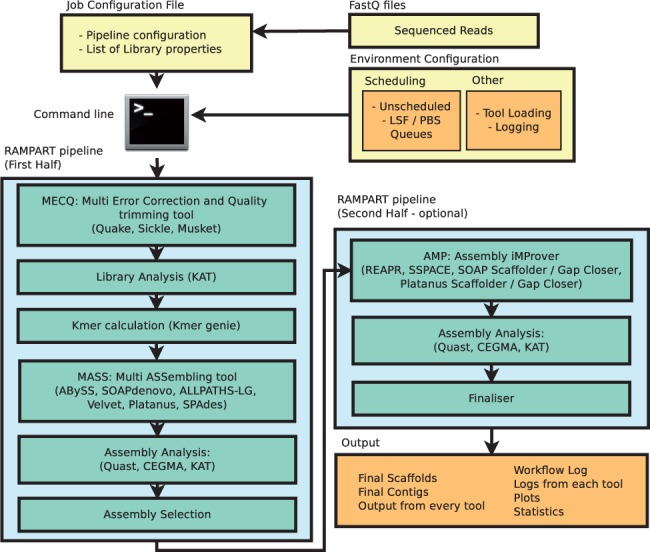

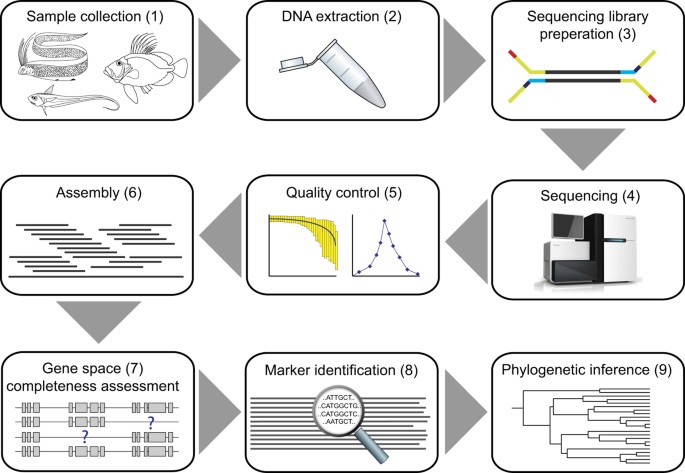



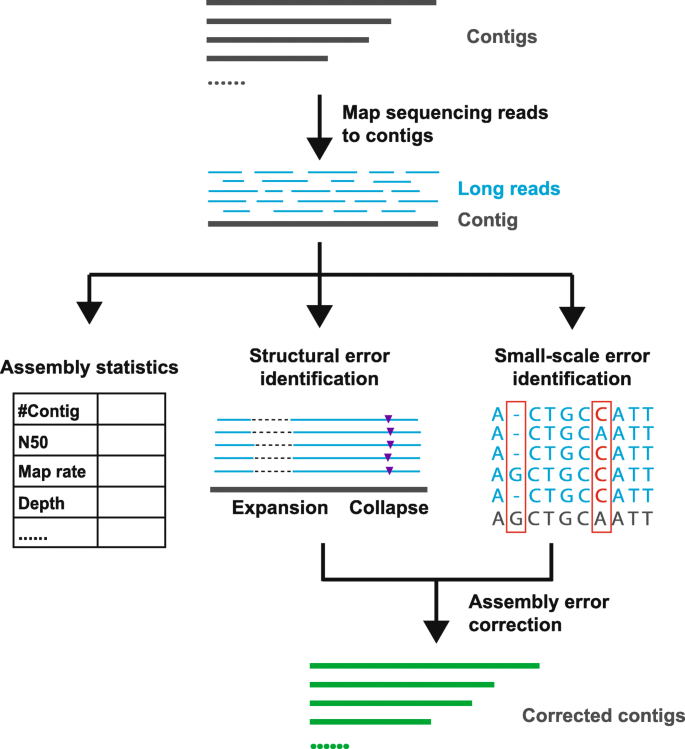
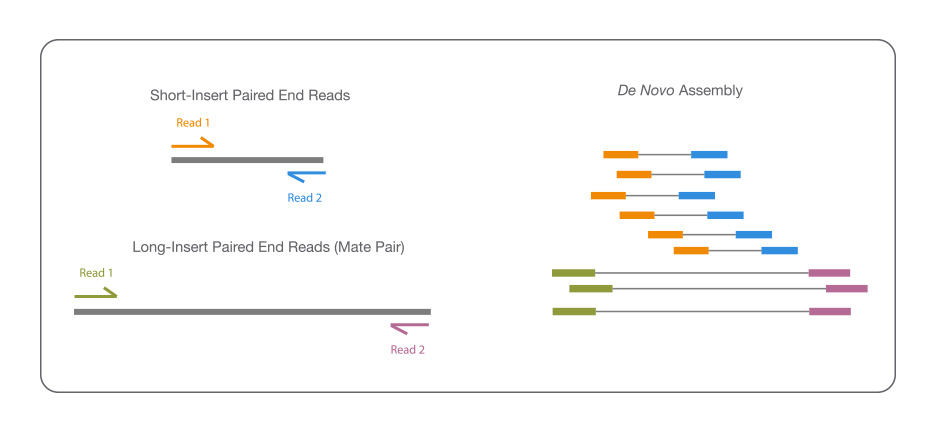
![PDF] RepARK—de novo creation of repeat libraries from whole-genome NGS reads | Semantic Scholar PDF] RepARK—de novo creation of repeat libraries from whole-genome NGS reads | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/b1a85a7249f556c25e875658fa5bd54bb5e1ae08/4-Figure1-1.png)